Novel multiomics technologies to decipher molecular signatures of infectious diseases
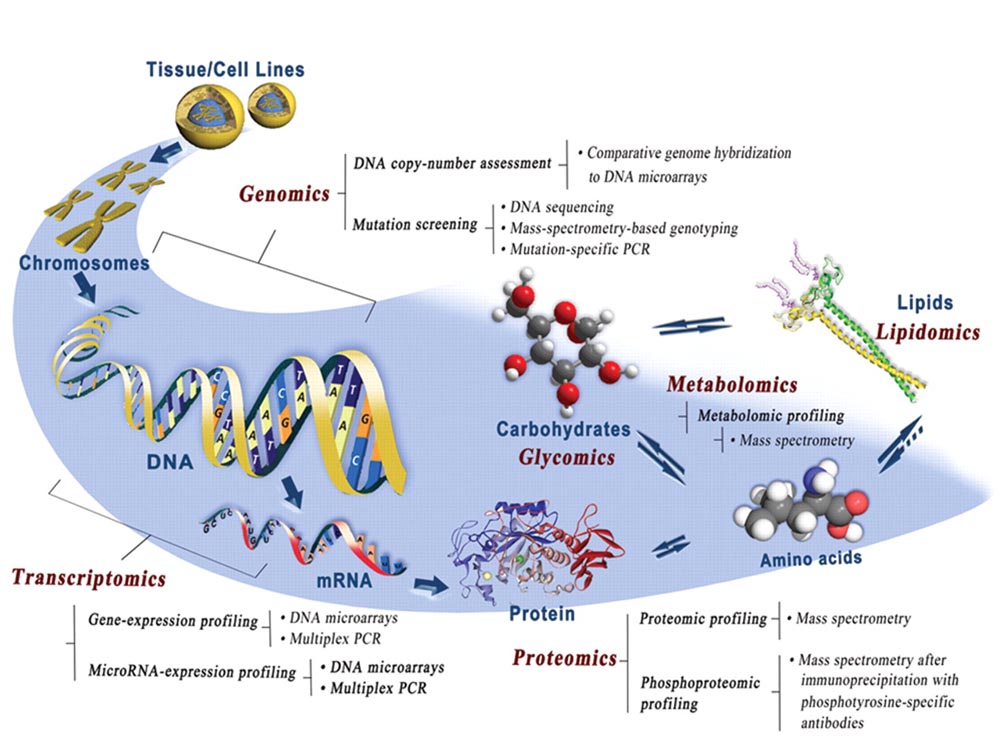
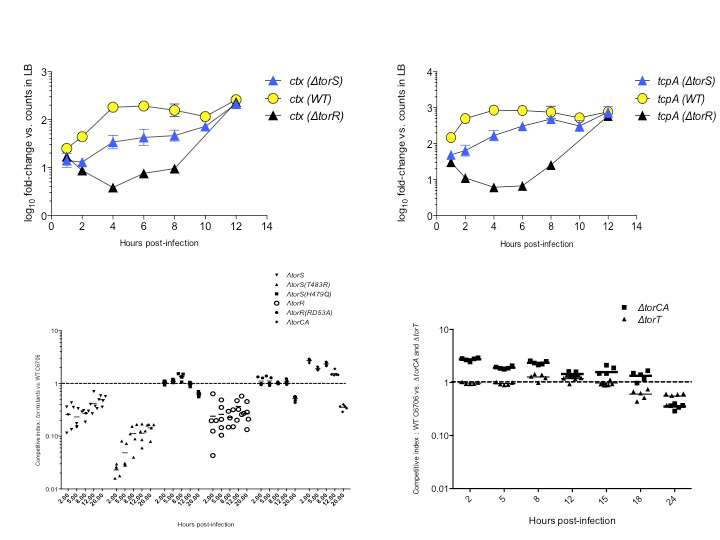
Signaling molecules, receptors, downstream regulators, and in vivo spatiotemporal expression kinetics for several virulence factors, and the subsequent host responses to infections are also not properly understood and defined. Using OMICS technologies such as NanoString assays and RNA-Seq, I uncovered the crucial contributions of bacterial dietary requirements to the capacity to cause diarrheal diseases. Two novel RNA and metabolic profiling approaches I have described can be ported into systems-biology platforms and possess a wide utility in microbiology research. Novel pathways and metabolites I have identified can serve as targets for prophylactic and therapeutic interventions against infectious diseases.
Peer-Reviewed Publications
-
New signaling pathways and metabolites for host: pathogen communication during gastrointestinal infections. 2014. [Abstract] [Poster] [Conference Proceedings]
Pratik Shah and Hung D
Proceedings of 5th American Society of Microbiology Conference on Cell-Cell Communication in Bacteria
-
Regulation of Vibrio cholerae virulence in response to novel host signal. 2012. [Poster] [Conference Proceedings]
Pratik Shah and Hung D
Gordon Research Conference on Microbial Toxins and Pathogenicity